Marine microbial eukaryotes form a foundational component of the evolutionary, food web and biogeochemical processes in the ocean. While these highly diverse organisms are typically 10-100 times less abundant than marine bacteria and archaea (and ten times further less abundant than marine viruses), they dramatically influence the diversity, ecology and evolution of nearly all marine microbial communities. Further, these microbes perform highly varied ecosystem functions, ranging from photosynthesis, heterotrophy and predation to parasitism. Only recently have studies emerged to evaluate the gene content and genomic attributes of these unique organisms to better understand their diversity and ecology. Such studies are only now possible with deep sequencing technology primarily due to the structurally complexity and extensive genome size of marine eukaryotic microbes (some of these genomes significantly dwarf the human genome).
The National Center for Genome Resources (NCGR), the Marine Microbiology Initiative (MMI) at the Gordon and Betty Moore Foundation and the international marine microbial eukaryote research community have recently completed a collaborative effort to sequence the transcriptomes (the expressed gene content from the genome) of more than 678 samples representing 210 unique genera, 305 species, and 409 strains. The effort provides a means to “fill in the blanks” on the eukaryotic evolutionary tree of cultured organisms, and rounds out MMI’s sequencing portfolio, which also contains the Microbial Genome Sequencing Project to sequence diverse marine bacterial and archaeal cultured isolates; the Marine Phage, Virus and Virome Sequencing Project to sequence viral isolates and viral metagenomes; and the Environmental Metagenomics Sequencing Portfolio to sequence whole microbial communities from locations around the world.
The MMETSP will enable researchers to deepen current understanding of microeukaryote ecology, nutritional strategies, evolution and roles in marine biogeochemistry. Moreover, this unique datasets will have far-reaching impact beyond the field of marine microbial ecology, providing a wealth of data which will transform current understanding of the basic biology of these organisms. Further information about the project is available through NCGR’s project webpage http://marinemicroeukaryotes.org/.
The related grant was awarded on September 29, 2010.
Bioinformatics Resource
A bioinformatics platform PhyloMETAREP (http://www.jcvi.org/phylo-metarep/) has been supported (Grant #2900; PI: Dr. Andy Allen) to enable exploration, visualization, and comparisons across annotated marine eukaryote transcriptomes. A key component of the PhyloMETAREP platform is a custom database, PhyloDB, which consists of manually annotated nuclear and organelle reference genomes and gene expression profiles suitable for genomic analyses of marine microeukaryotes and other microbes.
Data Availability
All data are publicly available as listed below, with accompanying accession information here. Datasets are also available through iMicrobe at the designated project page.
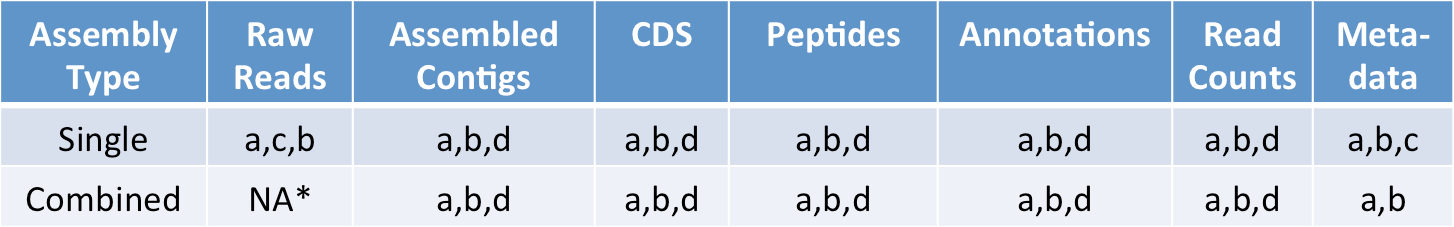
a. CAMERA http://camera.calit2.net/mmetsp/
b. iMicrobe http://data.imicrobe.us/project/view/104
c. SRA http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA231566
d. PhyloDB via PhyloMetarep† http://jcvi.org/metarep/
*The combined assemblies originate from reads from the single assemblies
†PhyloMetarep will have only one representation for each strain, either the combined assembly or a single assembly if only one sample was sequenced. Updated annotations have been completed and will be available soon. They include: BLAST against updated PhyloDB (including all MMETSP data) and KEGG and assignment of KEGG ortholog (KO) IDs, KOG, core eukaryotic gene annotation (CEGMA), Lineage Probability Index (LPI) assignment (to assist with filtering contaminants), transmembrane domain (tmhmm) and transporter annotation, pfam, tigrfam, superfamily, and GO. Combined assemblies have been integrated into a single annotation and read counts across conditions provided. Also, a 7-level taxonomy that is tied to PR2 has been implemented which will facilitate more meaningful comparative analyses at various levels of phylogenetic resolution. For questions, please contact Andy Allen (aallen@jcvi.org) or John McCrow (jmccrow@jcvi.org).
Contacts
For more information, please contact:
NCGR: Callum Bell (cjb@ncgr.org, +1 505-995-4428) or Stephanie Guida (sguida@ncgr.org)
GBMF: Jon Kaye (jon.kaye@moore.org, +1 650-213-3122)
iMicrobe: Bonnie Hurwitz (bhurwitz@email.arizona.edu) or Kenneth Youens-Clark (kyclark@email.arizona.edu)
For a list of participants in the bioinformatics internship program, please contact NCGR.
Publications as of May 2015
- Ryan, D.E., A. Pepper & L. Campbell. 2014. De novo assembly and characterization of the transcriptome of the toxic dinoflagellate Karenia brevis. BMC Genomics, 15, 888. doi:10.1186/1471-2164-15-888
- Keeling, P. J., F. Burki, H. M. Wilcox, B. Allam, E. E. Allen, L. A. Amaral-Zettler, E. V. Armbrust, J. M. Archibald, A. K. Bharti, C. J. Bell, B. Beszteri, K. D. Bidle, C. T. Cameron, L. Campbell, D. A. Caron, R. A. Cattolico, J. L. Collier, K. Coyne, S. K. Davy, P. Deschamps, S. T. Dyhrman, B. Edvardsen, R. D. Gates, C. J. Gobler, S. J. Greenwood, S. M. Guida, J. L. Jacobi, K. S. Jakobsen, E. R. James, B. Jenkins, U. John, M. D. Johnson, A. R. Juhl, A. Kamp, L. A. Katz, R. Kiene, A. Kudryavtsev, B. S. Leander, S. Lin, C. Lovejoy, D. Lynn, A. Marchetti, G. McManus, A. M. Nedelcu, S. Menden-Deuer, C. Miceli, T. Mock, M. Montresor, M. A. Moran, S. Murray, G. Nadathur, S. Nagai, P. B. Ngam, B. Palenik, J. Pawlowski, G. Petroni, G. Piganeau, M. C. Posewitz, K. Rengefors, G. Romano, M. E. Rumpho, T. Rynearson, K. B. Schilling, D. C. Schroeder, A. G. Simpson, C. H. Slamovits, D. R. Smith, G. J. Smith, S. R. Smith, H. M. Sosik, P. Stief, E. Theriot, S. N. Twary, P. E. Umale, D. Vaulot, B. Wawrik, G. L. Wheeler, W. H. Wilson, Y. Xu, A. Zingone, & A. Z. Worden. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol, 12(6), e1001889. doi: 10.1371/journal.pbio.1001889
- Rockwell, N. C., D. Duanmu, S. S. Martin, C. Bachy, D. C. Price, D. Bhattacharya, A. Z. Worden, & J. C. Lagarias. (2014). Eukaryotic algal phytochromes span the visible spectrum. Proc Natl Acad Sci U S A, 111(10), 3871-3876. doi: 10.1073/pnas.1401871111
- D'Adamo, S., R. E. Jinkerson, E. S. Boyd, S. L. Brown, B. K. Baxter, J. W. Peters, & M. C. Posewitz. (2014). Evolutionary and biotechnological implications of robust hydrogenase activity in halophilic strains of tetraselmis. PLoS One, 9(1), e85812. doi: 10.1371/journal.pone.0085812
- Koid, A., Z. Liu, R.Terrado, A. C. Jones, D. A. Caron, & K. B. Heidelberg. (2014). Comparative Transcriptome Analysis of Four Prymnesiophyte Algae. PLoS One, 9(6), e97801. doi: 10.1371/journal.pone.0097801.t001
- Imanian, B., & P. J. Keeling. (2014). Horizontal gene transfer and redundancy of tryptophan biosynthetic enzymes in dinotoms. Genome Biol Evol, 6(2), 333-343. doi: 10.1093/gbe/evu014
- Bender, S. J., C. A. Durkin, C. T. Berthiaume, R. L. Morales, & E. V. Armbrust. (2014). Transcriptional responses of three model diatoms to nitrate limitation of growth. Frontiers in Marine Science, 1. doi: 10.3389/fmars.2014.00003
- Tanifuji, G., N. T. Onodera, M. W. Brown, B. A. Curtis, A. J. Roger, G. Ka-Shu Wong, M. Melkonian, & J. M. Archibald. (2014). Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: convergent reductive evolution and frequent recombination in nucleomorph-bearing algae. BMC Genomics, 15, 374. doi: 10.1186/1471-2164-15-374
- Adelfi, M. G., M. Borra, R. Sanges, M. Montresor, A. Fontana, & M. I. Ferrante. (2014). Selection and validation of reference genes for qPCR analysis in the pennate diatoms Pseudo-nitzschia multistriata and P. arenysensis. Journal of Experimental Marine Biology and Ecology, 451, 74-81. doi: 10.1016/j.jembe.2013.11.003
- Santoferrara, L. F., S. Guida, H. Zhang, & G. B. McManus. (2014). De novo transcriptomes of a mixotrophic and a heterotrophic ciliate from marine plankton. PLoS One, 9(7), e101418. doi: 10.1371/journal.pone.0101418
- Grant, J. R., & L. A. Katz. (2014). Building a phylogenomic pipeline for the eukaryotic tree of life - addressing deep phylogenies with genome-scale data. PLoS Curr, 6. doi: 10.1371/currents.tol.c24b6054aebf3602748ac042ccc8f2e9
-
Frischkorn, K. R., M. J. Harke, C. J. Gobler, & S. T. Dyhrman. (2014). De novo assembly of Aureococcus anophagefferens transcriptomes reveals diverse responses to the low nutrient and low light conditions present during blooms. Front Microbiol, 5, 375. doi: 10.3389/fmicb.2014.00375
-
Rubin, E., E. P. Espinosa, A. Koller, & B. Allam. (2015). Characterisation of the secretome of the clam parasite, QPX. International Journal for Parasitology, 45(2-3): 187-196. doi:10.1016/j.ijpara.2014.10.008
-
Marchetti, A., D. Catlett, B. M. Hopkinson, K. Ellis, & N. Cassar. (2015). Marine diatom proteorhodopsins and their potential role in coping with low iron availability. ISME Journal. doi: 10.1038/ismej.2015.74
-
Alexander, H., B. D. Jenkins, T. A. Rynearson, & S. T. Dyhrman. (2015). Metatranscriptome analyses indicate resource partitioning between diatoms in the field. Proceedings of the National Academy of Sciences, 112(17): E2182-E2190. doi: 10.1073/pnas.1421993112
-
Curtis B. A., G. Tanifuji, F. Burki, A. Gruber, M. Irimia, S. Maruyama, M. C. Arias, S. G. Ball, G. H. Gile, Y. Hirakawa, J. F. Hopkins, A. Kuo, S. A. Rensing, J. Schmutz, A. Symeonidi, M. Elias, R. J. M. Eveleigh, E. K. Herman, M. J. Klute, T. Nakayama, M. Obornik, A. Reyes-Prieto, E. V. Armbrust, S. J. Aves, R. G. Beiko, P. Coutinho, J. B. Dacks, D. G. Durnford, N. M. Fast, B. R. Green, C. J. Grisdale, F. Hempel, B. Henrissat, M. P. Hoppner, K. I. Ishida, E. Kim, L. Koreny, P. G. Kroth, Y. Liu, S. B. Malik, U. G. Maier, D. McRose, T. Mock, J. A. D. Neilson, N. T. Onodera, A. M. Poole, E. J. Pritham, T. A. Richards, G. Rocap, S. W. Roy, C. Sarai, S. Schaack, S. Shirato, C. H. Slamovits, D. F. Spencer, S. Suzuki, A. Z. Worden, S. Zauner, K. Barry, C. Bell, A. K. Bharti, J. A. Crow, J. Grimwood, R. Kramer, E. Lindquist, S. Lucas, A. Salamov, G. I. McFadden, C. E. Lane, P. J. Keeling, M. W. Gray, I. V. Grigoriev, & J. M. Archibald. (2012). Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature, 492: 59-65. doi:10.1038/nature11681
-
Duanmu, D., C. Bachy, S. Sudek, C. H. Wong, V. Jimenez, N. C. Rockwell, S. S. Martin, C. Y. Ngan, E. N. Reistetter, M. J. van Baren, D. C. Price, C. L. Wei, A. Reyes-Prieto, J. C. Lagarias, & A. Z. Worden. (2014). Marine algae and land plants share conserved phytochrome signaling systems. Proceedings of the National Academy of Science, 111(44): 15827-15832. doi: 10.1073/pnas.1416751111
-
Mandel, M. J., M. S. Wollenberg, E. V. Stabb, K. L. Visick, & E. G. Ruby. (2009). A single regulatory gene is sufficient to alter bacterial host range. Nature, 458(7235): 215-218. doi: 10.1038/nature07660

Message sent
Thank you for sharing.